TABLE OF CONTENTS
- 1. ScPovPlot3D/Potential.inc
- 1.1. Potential.inc/CreateVFC
- 1.2. Potential.inc/CreateVFC2
- 1.3. Potential.inc/DeclareStructure
- 1.4. Potential.inc/Draw01CubicInterpolSrf
- 1.5. Potential.inc/DrawBones
- 1.6. Potential.inc/DrawBonesBW
- 1.7. Potential.inc/DrawCatRomSrf
- 1.8. Potential.inc/DrawKrigedSurf
- 1.9. Potential.inc/DrawKrigedSurf2
- 1.10. Potential.inc/DrawQVBox
- 1.11. Potential.inc/SetExLimiter, _exx
- 1.12. Potential.inc/SetGradMax
- 1.13. Potential.inc/SetGradMin
- 1.14. Potential.inc/SetGradMin2
- 1.15. Potential.inc/SetTanTau, _TanTau
- 1.16. Potential.inc/_K3a
- 1.17. Potential.inc/_K3b
- 1.18. Potential.inc/_K3den
ScPovPlot3D/Potential.inc [ Modules ]
PURPOSE
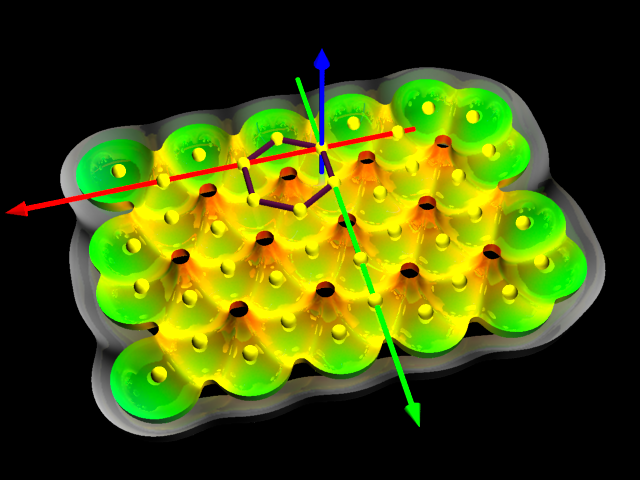
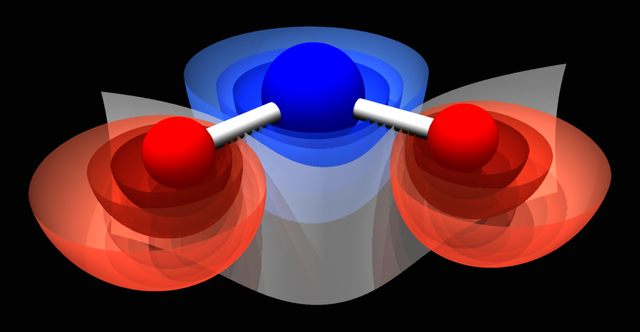
3D visualization of scalar fields (Coulomb & Lenard-Jones, grid defined. Partially done - any externally calculated on the grid.) using isosurfaces & other techniques.
Fig.[Potential] Examples of visualisation of coulombean potential around the water molecule
VERSION
4.0.0, tested on PovRay 3.7.
AUTHOR
Janusz Opiła Ph.D.
jmo{at}agh.edu.pl, janusz.opila{at}gmail.com
Dept. of Applied Informatics,
https://www.facebook.com/KatedraInformatykiStosowanejWZAGH/
http://kis.zarz.agh.edu.pl/
AGH University of Science & Technology, Cracow, Poland
Maintained by Janusz Opiła Ph.D.
Homepage: http://scpovplot3d.sourceforge.net
HISTORY
2016-01-05 - Tricubic interpolation, KDE interpolation 2016-02-03 - inserted procedures for isosurface interpolation
COPYRIGHT
GNU GPL v.3 License (c) 2012-now by Janusz Opiła Ph.D. AGH University of Science and Technology
Potential.inc/CreateVFC [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Coulombean potential from network of electric charges. Results are computed in [V], for distance given in Angstrems, and charge given in 'e' units.
SYNOPSIS
#macro CreateVFC(_PTs) //
INPUTS
garray - array defining structure, see Structures.inc
OUTPUTS
Function
SEE ALSO
CreateVFC2, CreateVLJ, CreateVLJ2
Potential.inc/CreateVFC2 [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Coulombean potential from network of electric charges. Results are computed in [V], for distance given in Angstrems, and charge given in 'e' units.
SYNOPSIS
#macro CreateVFC2(_PTs, _FF) // Coulombean potential from network of electric charges
INPUTS
garray - array defining structure, see Structures.inc float - unit divider, ex.: use 1000 for [mV], and 0.001 for [kV]. Volatile setting, may be changed in future releases.
OUTPUTS
Function
SEE ALSO
CreateVFC, CreateVLJ, CreateVLJ2
Potential.inc/Draw01CubicInterpolSrf [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Computes isosurface usin TriCubic interpolation. Setting variable "Debug" to more than zero produces diagnostic output
SYNOPSIS
#macro Draw01CubicInterpolSrf (_trsh, _dd) // _trsh - isosurface threshold
INPUTS
_trsh - double - threshold value of isosurface _dd - double - edge length of elementary cell QVr[] - garray - implicit attribute, nodes of computation frame
OUTPUTS
None,
SIDE EFFECTS
None or unknown
SEE ALSO
DrawCubicInterpolSrf ()
Potential.inc/DrawBones [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Draws all 64 nodes used by interpolation procedures. For illustration purposes only.
SYNOPSIS
#macro DrawBones (_dd, _xyz) // default look
INPUTS
_dd - double - distance parameter, _dd > 0. _xyz - 3Dvector - additional shift of the whole structure
OUTPUTS
none
SEE ALSO
DrawQVBox (), DrawAllCells (), DrawBonesBW ()
Potential.inc/DrawBonesBW [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Draws all 64 nodes used by interpolation procedures. For illustration purposes only.
SYNOPSIS
#macro DrawBonesBW (_dd, _xyz) // default look
INPUTS
_dd - double - distance parameter, _dd > 0. _xyz - 3Dvector - additional shift of the whole structure
OUTPUTS
none
SEE ALSO
DrawQVBox (), DrawAllCells (), DrawBonesBW ()
Potential.inc/DrawCatRomSrf [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Computes isosurface usin TriCubic interpolation. Setting variable "Debug" to more than zero produces diagnostic output
SYNOPSIS
#macro DrawCatRomSrf (_trsh, _dd) // _trsh - isosurface threshold
INPUTS
_trsh - double - threshold value of isosurface _dd - double - edge length of elementary cell QVr[] - garray - implicit attribute, nodes of computation frame
OUTPUTS
None,
SIDE EFFECTS
None or unknown
SEE ALSO
DrawCubicInterpolSrf (), Draw01CubicInterpolSrf ()
Potential.inc/DrawKrigedSurf [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Computes isosurface usin DrawKrigeSurf interpolation. Setting variable "Debug" to more than zero produces diagnostic output
SYNOPSIS
#macro DrawKrigedSurf (_trsh, _dd, _pp) // __1
INPUTS
_trsh - double - threshold value of isosurface _dd - double - edge length of elementary cell _pp - smoothing constant QVr[] - garray - implicit attribute, nodes of computation frame
OUTPUTS
None,
SIDE EFFECTS
None or unknown
SEE ALSO
DrawCubicInterpolSrf (), Draw01CubicInterpolSrf (), DrawKrigedSurf2 ()
Potential.inc/DrawKrigedSurf2 [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Computes isosurface using gaussian KDE interpolation. Setting variable "Debug" to more than zero produces diagnostic output Features full frame (ie. all 64 neighbours) in reduced coordinates.
SYNOPSIS
#macro DrawKrigedSurf2 (_trsh, _dd, _pp) // __1
INPUTS
_trsh - double - threshold value of isosurface _dd - double - edge length of elementary cell _pp - smoothing constant QVr[] - garray - implicit attribute, nodes of computation frame
OUTPUTS
None,
SIDE EFFECTS
None or unknown
SEE ALSO
DrawCubicInterpolSrf (), Draw01CubicInterpolSrf (), DrawKrigedSurf ()
Potential.inc/DrawQVBox [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Draws box containing computed grid of values
SYNOPSIS
#macro DrawQVBox(_QVr) // default look
INPUTS
_QVr - garray
OUTPUTS
none
SEE ALSO
DrawBones (), DrawAllCells ()
Potential.inc/SetExLimiter, _exx [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Parameter for elementary cell selector. Default '0'. The greater the larger tolerance. For Catmul-Rom values >0 impose risk of artifacts.
SYNOPSIS
#ifndef (_exx) #declare _exx = 0.0; #end #macro SetExxLimiter(_sx)
INPUTS
_sx - double - parameter value
OUTPUTS
sets parameter
SEE ALSO
DrawCatRomSrf (), crSm0 (), crS12 ()
Potential.inc/SetGradMax [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Sets parameter _gMin for gradient texture mapping
SYNOPSIS
#macro SetGradMax(_g)
INPUTS
_g - double - parameter value
OUTPUTS
sets internal parameter _gMin2
SEE ALSO
SetGradMin (), SetGradMin2 ()
Potential.inc/SetGradMin [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Sets parameter _gMin for gradient texture mapping
SYNOPSIS
#macro SetGradMin(_g)
INPUTS
_g - double - parameter value
OUTPUTS
sets internal parameter _gMin
SEE ALSO
SetGradMin2 (), SetGradMax ()
Potential.inc/SetGradMin2 [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Sets parameter _gMin for gradient texture mapping
SYNOPSIS
#macro SetGradMin2(_g)
INPUTS
_g - double - parameter value
OUTPUTS
sets internal parameter _gMin2
SEE ALSO
SetGradMin (), SetGradMax ()
Potential.inc/SetTanTau, _TanTau [ Main macros ]
[ Top ] [ Potential.inc ] [ Main macros ]
PURPOSE
Parameter for tangent calculations in Catmul-Rom formulation
SYNOPSIS
#ifndef (_TanTau) #declare _TanTau = 0.5; #end #macro SetTanTau(_tt)
INPUTS
_tt - double - parameter value
OUTPUTS
sets parameter
SEE ALSO
DrawCatRomSrf (), crSm0 (), crS12 ()
Potential.inc/_K3a [ Helper macros ]
[ Top ] [ Potential.inc ] [ Helper macros ]
PURPOSE
Computes KDE approximation using gaussian interpolation. Whole interpolation is split in three functions, _K3a, _K3b, _K3den due to limitation imposed on number of arguments. Total output equals: [(_K3a()+_K3b())/_K3den()] - denominator is computed in _K3den () fucnction, Model is computed in reduced space, ie. rx,ry,rz is assumed zero. Setting variable "Debug" to more than zero produces diagnostic output.
SYNOPSIS
#declare _K3a = Function( // Full cube core nodes, internal part 1; __2 xx, yy, zz // xP, yP, zP: float - internally supplied ,_d, __d // edge length/ double length of elementary cell ,_p // smoothing constant ,_Q000, _Q010, _Q100, _Q110 // lower base ,_Q001, _Q011, _Q101, _Q111 // upper base ,_Qm00, _Qm10, _Q200, _Q210 // X l ,_Qm01, _Qm11, _Q201, _Q211 // X u ,_Q0m0, _Q020, _Q1m0, _Q120 // Y l ,_Q0m1, _Q021, _Q1m1, _Q121 // Y u ,_Q00m, _Q01m, _Q10m, _Q11m // Z l ,_Q002, _Q012, _Q102, _Q112 // Z u )
INPUTS
xx ,yy, zz - double - supplied by POV-Ray, current point in space
_d, __d - double - edge length/ double length of elementary cell
_p - double - smoothing constant,
Quvw - double - values in nodes. uvw->{m,0,1,2}: m:=i-1, 0:=i, 1:=i+1, 2:=i+2
OUTPUTS
None,
SIDE EFFECTS
None or unknown
SEE ALSO
DrawCubicInterpolSrf (), Draw01CubicInterpolSrf (), _K3b
Potential.inc/_K3b [ Helper macros ]
[ Top ] [ Potential.inc ] [ Helper macros ]
PURPOSE
Computes KDE approximation using gaussian interpolation. Whole interpolation is split in three functions, _K3a, _K3b, _K3den due to limitation imposed on number of arguments. Total output equals: [(_K3a()+_K3b())/_K3den()] - denominator is computed in _K3dem () fucnction, Model is computed in reduced space, ie. rx,ry,rz is assumed zero. Setting variable "Debug" to more than zero produces diagnostic output.
SYNOPSIS
#declare _K3b = Function( // Full cube core nodes, internal part 1; __2 xx, yy, zz // xP, yP, zP: float - internally supplied ,_d, __d // edge length/ double length of elementary cell ,_p // smoothing constant ,_Qmmm, _Q0mm, _Q1mm, _Q2mm // lower dozen __m ,_Qm0m, _Q20m // lower dozen __m ,_Qm1m, _Q21m // lower dozen __m ,_Qm2m, _Q02m, _Q12m, _Q22m // lower dozen __m ,_Qmm2, _Q0m2, _Q1m2, _Q2m2 // upper dozen __2 ,_Qm02, _Q202 // upper dozen __2 ,_Qm12, _Q212 // upper dozen __2 ,_Qm22, _Q022, _Q122, _Q222 // upper dozen __2 ,_Qmm0, _Q2m0, _Q220, _Qm20 // 0 level fourth __0 ,_Qmm1, _Q2m1, _Q221, _Qm21 // 1 level fourth __1 )
INPUTS
xx ,yy, zz - double - supplied by POV-Ray, current point in space
_d, __d - double - edge length/ double length of elementary cell
_p - double - smoothing constant,
Quvw - double - values in nodes. uvw->{m,0,1,2}: m:=i-1, 0:=i, 1:=i+1, 2:=i+2
OUTPUTS
None,
SIDE EFFECTS
None or unknown
SEE ALSO
DrawCubicInterpolSrf (), Draw01CubicInterpolSrf (), _K3b
Potential.inc/_K3den [ Helper macros ]
[ Top ] [ Potential.inc ] [ Helper macros ]
PURPOSE
Computes KDE approximation using gaussian interpolation. Whole interpolation is split in three functions, _K3a, _K3b, _K3den due to limitation imposed on number of arguments. Total output equals: [(_K3a()+_K3b())/_K3den()] - denominator is computed in _K3den () Function, Model is computed in reduced space, ie. rx,ry,rz is assumed zero. Setting variable "Debug" to more than zero produces diagnostic output.
SYNOPSIS
#declare _K3den = Function( // Full cube core nodes, internal part 1; __2 xx, yy, zz // xP, yP, zP: float - internally supplied ,_d, __d // edge length/ double length of elementary cell ,_p // smoothing constant )
INPUTS
xx, yy, zz - double - supplied by POV-Ray, current point in space _d, __d - double - edge length/ double length of elementary cell _p - double - smoothing constant, Quvw - double - values in nodes. uvw->\m,0,1,2\: m:=i-1, 0:=i, 1:=i+1, 2:=i+2
OUTPUTS
None,
SIDE EFFECTS
None or unknown
SEE ALSO
DrawCubicInterpolSrf (), Draw01CubicInterpolSrf (), _K3a (), _K3b ()